Mass Spectrometry-Based Proteomic Evidence: A Cornerstone in Modern Biological Research
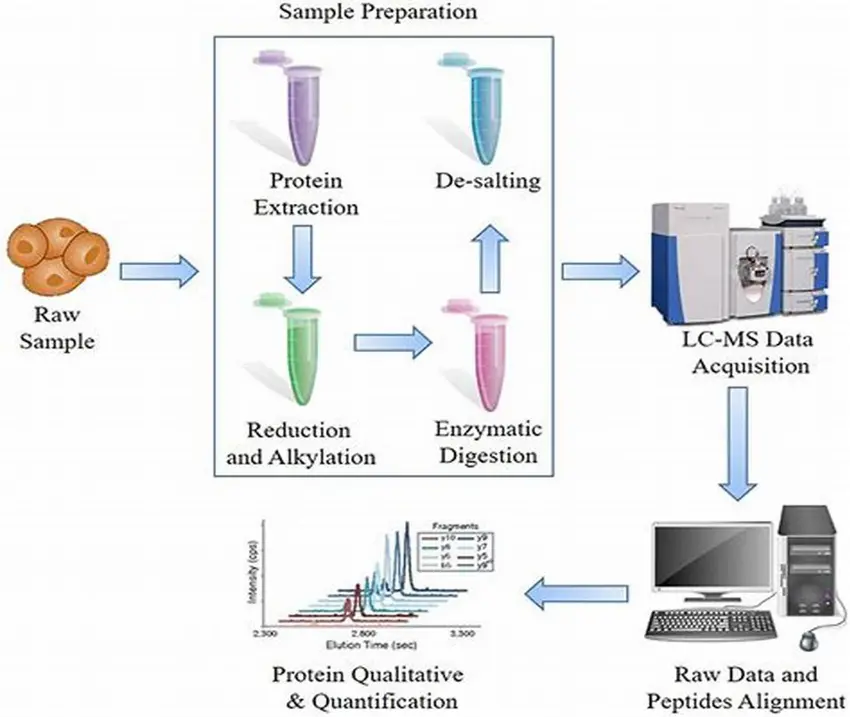
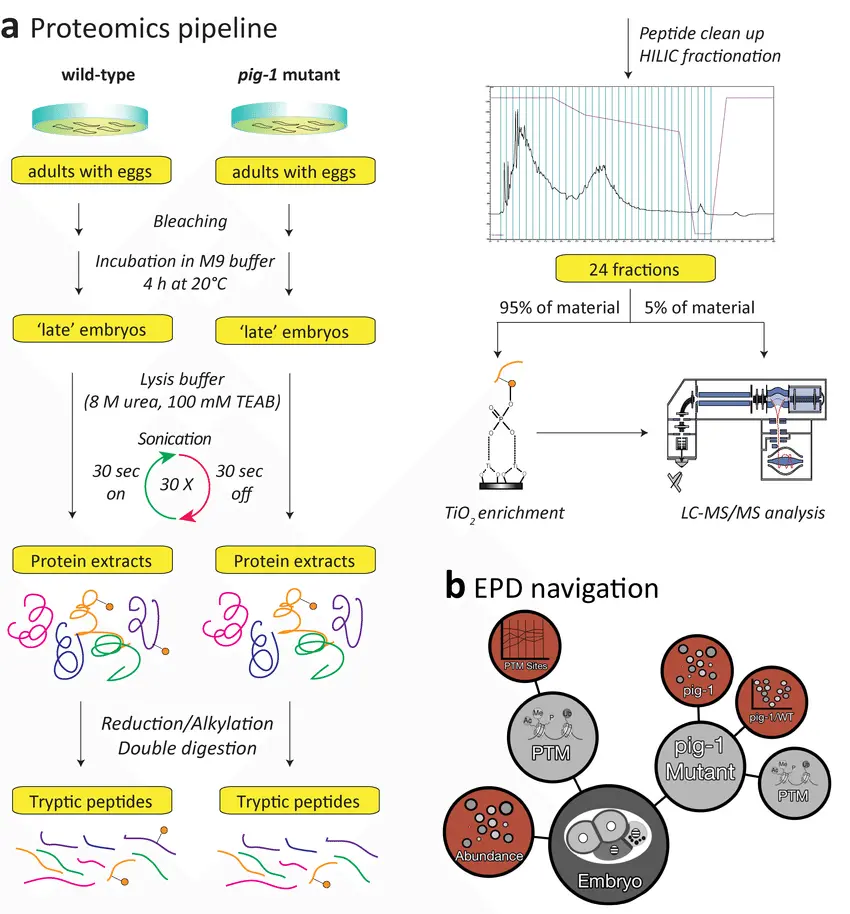
Mass spectrometry (MS) has become one of the most powerful tools in modern biology, enabling scientists to gain an unprecedented view of the proteome the complete set of proteins expressed by a cell, tissue, or organism. Unlike genomics or transcriptomics, which provide indirect information on potential protein expression, MS-based proteomics delivers direct molecular evidence of proteins, their isoforms, and post-translational modifications. This makes it indispensable for refining genome annotations, validating predicted proteins, and discovering novel biological pathways.
At its core, MS-based proteomic evidence is generated by identifying and quantifying peptides derived from protein digestion. These peptides are separated by liquid chromatography (LC) and analyzed with high-resolution MS instruments, producing spectra that can be matched to protein databases or de novo reconstructed. Such approaches reveal not only the presence of canonical proteins but also unexpected variants, including alternative splice products, single amino acid polymorphisms, and proteoforms that are invisible to DNA- or RNA-based methods.
The impact of MS-based proteomics spans across multiple fields

Biomedical research
MS helps to uncover disease-specific protein markers, supporting early diagnosis and targeted therapy.

Proteogenomics
By integrating proteomic data with genomic and transcriptomic information, researchers can refine gene models and validate predicted coding regions.

Systems biology
Quantitative proteomics allows the mapping of complex protein networks and signaling pathways, advancing our understanding of cellular mechanisms.