Why ProteoAnnotator is Essential for Your Research
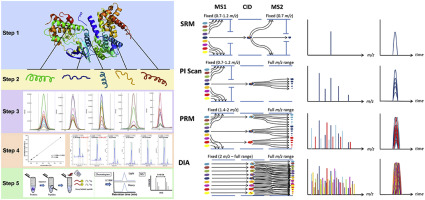
Mass spectrometry (MS) data is vital in analyzing proteins and genomes. ProteoAnnotator streamlines the process of integrating MS spectra with genomic data. Using an automated two-step pipeline, it allows you to:
- Verify that predicted genes are translated into proteins.
- Accurately identify peptides from your samples.
- Filter out irrelevant spectra to focus on the key results.
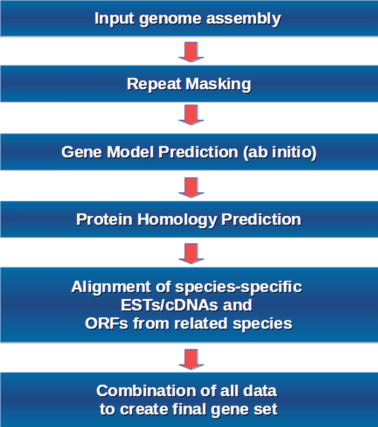
Main Genome Annotation Steps
Genome annotation involves several key steps, such as repeat masking, protein homology prediction, and the alignment of open reading frames (ORFs) from other species. These steps often depend on data from previous genome assemblies and annotations. If these earlier annotations contain errors, those mistakes can be carried over, leading to reduced accuracy in the final annotation. This transfer of errors can affect the overall precision, making it important to carefully validate each step with reliable data to ensure better accuracy.
Applications in Various Research Fields
1. Genomic Research
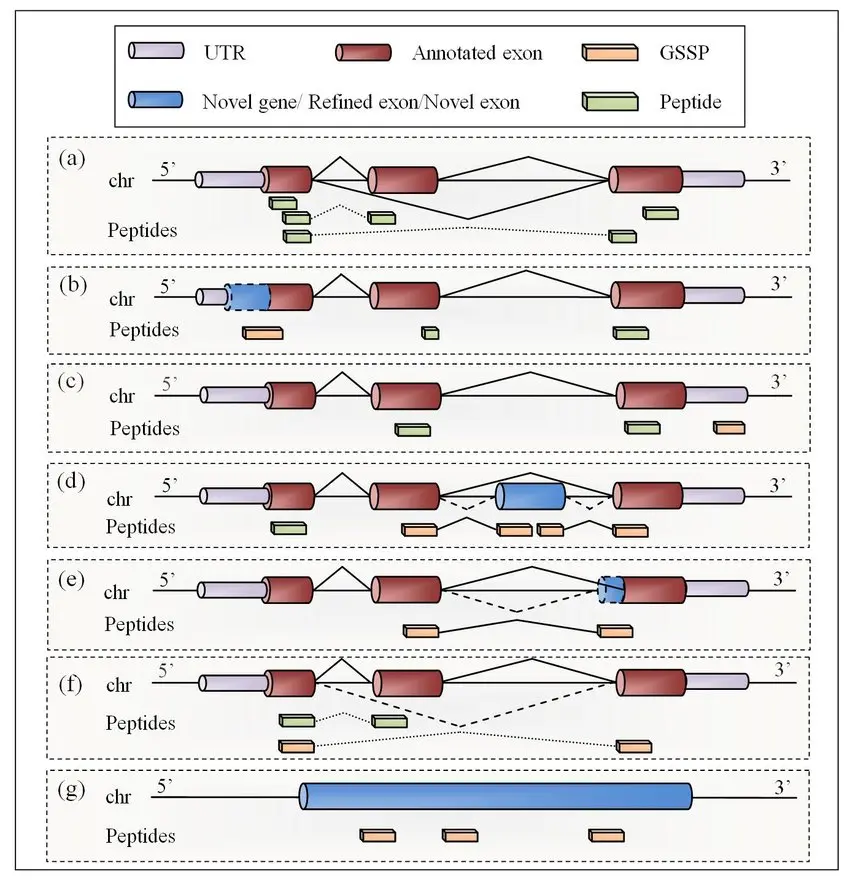
ProteoAnnotator supports genomic research by validating whether predicted gene sequences are accurately expressed as proteins. This process is vital for ensuring the accuracy of genome annotations, which are essential for downstream analyses such as gene function identification and the study of genomic variability. By confirming gene-to-protein translation through mass spectrometry data, ProteoAnnotator helps researchers refine their genomic models and improve the overall quality of the genome sequence.
2. Biotechnology
In biotechnology, ProteoAnnotator plays a crucial role in optimizing recombinant protein production. It aids in ensuring that the proteins produced in biological systems match the expected peptide sequences, which is key for quality control in biomanufacturing. The tool is also useful for identifying new protein targets, whether for therapeutic applications or for understanding the molecular mechanisms underlying biological processes. The ability to identify and accurately map peptides enhances the development of novel biotechnological applications, from enzyme production to biosensors.
3. Proteomics
Proteomics research involves the large-scale study of proteins, which requires highly accurate identification and characterization of proteins in complex biological samples. ProteoAnnotator simplifies this process by filtering out irrelevant mass spectrometry data, allowing researchers to focus on the most relevant peptides and proteins. It helps ensure high-quality results in protein identification, including the detection of post-translational modifications (PTMs), which are essential for understanding protein functionality, interaction networks, and cellular pathways.
Start Using ProteoAnnotator Today
Convert your MS data into useful insights with ProteoAnnotator. Simplify your workflow, improve result accuracy, and speed up your research.